Since sequences in the background set used for comparison also contain RCGTG motifs, this enrichment likely arises from the well known preference for A versus G in the first position of the HIF Orbifloxacin binding consensus. These results collectively suggest that several additional transcription factors could influence HIF transcriptional activity. Importantly, we noted that most of the enriched TFBSs corresponded to stress-responsive transcription factors. Varied stress-responsive TFs have been shown to coordinately regulate the same genes, 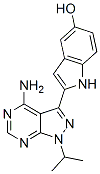 and indeed several transcription factors are activated by the same stresses in mammalian cells. However, it is unclear whether this cooperation among stressresponsive pathways translates at the genomic level. In order to evaluate the functional significance of the TFBSs enriched in core HIF binding regions, we carried out an experimental validation by disrupting selected sequences in bona fide HIF-responsive promoters. Importantly, no experimental confirmation had been attempted on previously reported predictions, and therefore the biological significance of those findings remained unclear. In spite of being limited to three selected promoters, our results clearly indicate that, compared to control mutations, alteration of binding sequences of transcription factors enriched in HIF binding regions, and different from HIFs themselves, have a specific effect on the transcriptional activation of HIF-responsive promoters. In particular, we found negative effects on hypoxic induction of LDHA and GYS1 promoters upon disruption of CREB and CEBPB binding sites proximal to the HRE, whereas mutation of an AP1 site proximal to the CA9 HRE led to a slightly augmented hypoxic induction of the promoter. In agreement with our results, mutation of the same CREB binding site was been previously shown to alter LDHA hypoxic induction. Interestingly, USF binding to a palindrome CACGTG HRE in the LDHA promoter was suggested to complement HIF binding. However, our results do not allow us to corroborate these findings, as mutation of this HRE was not evaluated in our experiments. Furthermore, hypoxic CA9 Catharanthine sulfate expression has been linked to cooperation between AP1 family member ATF4 and HIF1a. In this study, ATF4 overexpression led to an augmented CA9 induction in hypoxia, with reduced hypoxic expression of CA9 being observed upon ATF4 knock-down. Chromatin immunoprecipitation experiments mapped ATF4 binding to the 21400/21000 region of the CA9 promoter, which falls outside of the promoter region employed in our experiments. Nevertheless, the apparent paradox with our results argues for careful interpretation of the role of AP1 in the HIF transcriptional response. In fact, both positive and negative effects of AP1 have been reported on hypoxic gene expression and, given the number of AP1 family members, these probably arise from compositional differences in AP1 complexes. Importantly, the effects observed upon mutation of CREB, CEBPB or AP1 binding sites were always moderate when compared to mutation of the HIF binding consensus RCGTG, suggesting that rather than being an absolute requirement for hypoxic induction, the integrity of these neighboring TFBSs fine-tunes the HIF-mediated transcriptional response. Thus, it is possible that multiple independent factors contribute, in an additive fashion, to HIF-mediated transcription. This model could also explain why we found a relatively large number of enriched TFBSs in HIF binding regions, but all of them sharing a modest statistical significance. On the whole, these observations indicate that several of the enriched TFBSs identified in our approach are of functional relevance for HIF-mediated transcription. Nevertheless, it should be noted that other TFs for which collaboration with HIFs has been previously suggested are not recovered as enriched in our approach.
and indeed several transcription factors are activated by the same stresses in mammalian cells. However, it is unclear whether this cooperation among stressresponsive pathways translates at the genomic level. In order to evaluate the functional significance of the TFBSs enriched in core HIF binding regions, we carried out an experimental validation by disrupting selected sequences in bona fide HIF-responsive promoters. Importantly, no experimental confirmation had been attempted on previously reported predictions, and therefore the biological significance of those findings remained unclear. In spite of being limited to three selected promoters, our results clearly indicate that, compared to control mutations, alteration of binding sequences of transcription factors enriched in HIF binding regions, and different from HIFs themselves, have a specific effect on the transcriptional activation of HIF-responsive promoters. In particular, we found negative effects on hypoxic induction of LDHA and GYS1 promoters upon disruption of CREB and CEBPB binding sites proximal to the HRE, whereas mutation of an AP1 site proximal to the CA9 HRE led to a slightly augmented hypoxic induction of the promoter. In agreement with our results, mutation of the same CREB binding site was been previously shown to alter LDHA hypoxic induction. Interestingly, USF binding to a palindrome CACGTG HRE in the LDHA promoter was suggested to complement HIF binding. However, our results do not allow us to corroborate these findings, as mutation of this HRE was not evaluated in our experiments. Furthermore, hypoxic CA9 Catharanthine sulfate expression has been linked to cooperation between AP1 family member ATF4 and HIF1a. In this study, ATF4 overexpression led to an augmented CA9 induction in hypoxia, with reduced hypoxic expression of CA9 being observed upon ATF4 knock-down. Chromatin immunoprecipitation experiments mapped ATF4 binding to the 21400/21000 region of the CA9 promoter, which falls outside of the promoter region employed in our experiments. Nevertheless, the apparent paradox with our results argues for careful interpretation of the role of AP1 in the HIF transcriptional response. In fact, both positive and negative effects of AP1 have been reported on hypoxic gene expression and, given the number of AP1 family members, these probably arise from compositional differences in AP1 complexes. Importantly, the effects observed upon mutation of CREB, CEBPB or AP1 binding sites were always moderate when compared to mutation of the HIF binding consensus RCGTG, suggesting that rather than being an absolute requirement for hypoxic induction, the integrity of these neighboring TFBSs fine-tunes the HIF-mediated transcriptional response. Thus, it is possible that multiple independent factors contribute, in an additive fashion, to HIF-mediated transcription. This model could also explain why we found a relatively large number of enriched TFBSs in HIF binding regions, but all of them sharing a modest statistical significance. On the whole, these observations indicate that several of the enriched TFBSs identified in our approach are of functional relevance for HIF-mediated transcription. Nevertheless, it should be noted that other TFs for which collaboration with HIFs has been previously suggested are not recovered as enriched in our approach.